Two researchers from Skoltech and McGill University have published a review of the promising research field known as structural proteomics in the influential journal Chemical Reviews. This field combines protein chemistry and mass spectrometry, an advanced analytical technique for identifying the chemical composition of substances based on precise mass. The ultimate goal is to resolve the detailed structure of proteins and therefore understand pathological processes, like Alzheimer’s, on the molecular level and predict drug candidates against diverse diseases faster and more efficiently.
Structural proteomics is concerned with identifying the structure of interacting proteins and the sites on proteins that interact with other proteins, including antibodies, or with drug molecules. This is made possible by a range of techniques providing complementary structural data: surface modification, limited proteolysis, hydrogen-deuterium exchange, and cross-linking.
“Consider a situation with serious health implications, where two proteins are fused together. Sometimes you need to prevent this sticking from happening. For example, this process is thought to be behind certain cancers,” study co-author and Skoltech Visiting Professor Christoph Borchers commented.
Besides determining protein interaction sites, another important aspect of structural proteomics is exploring the effects of so-called conformation changes. This refers to proteins adopting different spatial configurations without any changes to their chemical makeup: simply by folding into shapes. Researchers study conformation changes because they affect what interaction sites on proteins are actually exposed and therefore available to drug molecules or other proteins.
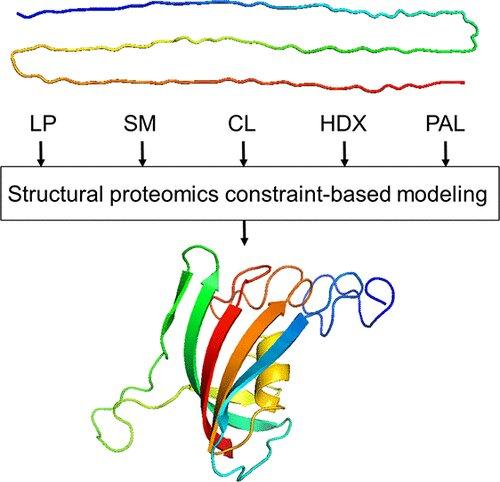
“Say, you have an ‘unhealthy’ conformation change for some reason,” Borchers said. “All of a sudden you’ve got an interaction going on, and the next thing you know — protein starts aggregating and there’s a fiber plaque forming — this is what happens in Alzheimer’s and Parkinson’s diseases. We need to know what this pathological conformation change is, along with the structure of the resulting aggregate.”
“Artificial intelligence is of much consequence here. While it cannot explain everything, it certainly adds a lot. We can go a long way in terms of understanding conformation changes and their role in neurodegenerative diseases, for example — by combining protein chemistry, molecular modeling, and AI,” study co-author Evgeniy Petrotchenko added, citing this as possibly the one biggest application of structural proteomics.
“Because sure, people are getting older, but they are still getting either cancer, or neurodegenerative diseases, or CVD [cardiovascular diseases],” Borchers continued, noting that 90% of all new cancer drugs are antibodies — and structural proteomics can determine protein-antibody interactions, too.
“It’s a very exciting field to be in,” the researcher said. “We’ve been doing this for almost 30 years now, and we’ve made great progress developing software packages for deciphering dynamic changes that elude conventional structural analysis tools, such as X-ray crystallography, nuclear magnetic resonance, or electron microscopy. And we’ve developed a range of complementary approaches: cross-linking studies, hydrogen-deuterium exchange, etc.”
Another thing about structural proteomics is the sheer scale it operates on. Imagine there was a drug whose mode of action you needed to characterize: Where exactly is it binding the target protein? (Proteins are, in fact, the molecules targeted by the vast majority of drugs.) The function of a protein is defined by its structure, and structural proteomics can determine the structure not just of one protein but of many, involved in a simultaneous interaction. “We are looking at hundreds of protein-protein interactions at a time. We are looking at the entire protein network. This cannot be done by other techniques,” Borchers explained. “Same as with an orchestra: You have to consider the whole thing.”
Reference: Petrotchenko EV, Borchers CH. Protein chemistry combined with mass spectrometry for protein structure determination. Chem Rev. 2021. doi: 10.1021/acs.chemrev.1c00302
This article has been republished from the following materials. Note: material may have been edited for length and content. For further information, please contact the cited source.









