Significance
Detection of molecular interactions is the foundation for many important biotechnology applications in society and industry, such as drug discovery, diagnostics, and DNA sequencing. This report describes a broadly applicable platform for detecting molecular interactions at the single-molecule scale, in real-time, label-free, and potentially highly multiplexable fashion, using single-molecule sensors on a highly scalable semiconductor sensor array chip. Such chips are both practically manufacturable in the near term, and have a durable long-term scaling roadmap, thus providing an ideal way to bring the power of modern chip technology to the broad area of biosensing. This work also realizes a 50-year-old scientific vision of integrating single molecules into electronic chips to achieve the ultimate miniaturization of electronics.
Abstract
For nearly 50 years, the vision of using single molecules in circuits has been seen as providing the ultimate miniaturization of electronic chips. An advanced example of such a molecular electronics chip is presented here, with the important distinction that the molecular circuit elements play the role of general-purpose single-molecule sensors. The device consists of a semiconductor chip with a scalable array architecture. Each array element contains a synthetic molecular wire assembled to span nanoelectrodes in a current monitoring circuit. A central conjugation site is used to attach a single probe molecule that defines the target of the sensor. The chip digitizes the resulting picoamp-scale current-versus-time readout from each sensor element of the array at a rate of 1,000 frames per second. This provides detailed electrical signatures of the single-molecule interactions between the probe and targets present in a solution-phase test sample. This platform is used to measure the interaction kinetics of single molecules, without the use of labels, in a massively parallel fashion. To demonstrate broad applicability, examples are shown for probe molecule binding, including DNA oligos, aptamers, antibodies, and antigens, and the activity of enzymes relevant to diagnostics and sequencing, including a CRISPR/Cas enzyme binding a target DNA, and a DNA polymerase enzyme incorporating nucleotides as it copies a DNA template. All of these applications are accomplished with high sensitivity and resolution, on a manufacturable, scalable, all-electronic semiconductor chip device, thereby bringing the power of modern chips to these diverse areas of biosensing.
Rapid, specific, and sensitive measurements of target analytes are the goals of many methods used in molecular biology and biotechnology. Bulk methods typically use a binding molecule to recognize the target molecule, combined with indirect optical reporter mechanisms, such as fluorescent dye labels or changes in bulk optical properties resulting from target binding. Such classical methods detect an average over many molecular binding events, and over timescales much longer than that of the primary molecular interactions. In contrast, the binding interactions of interest fundamentally occur at the single-molecule level, and are typically dynamic and stochastic in time, and thus contain far more detail than what is reflected in bulk reaction rates. This highlights the potential to access a fundamentally richer and more powerful level of information when measuring molecular interactions.
Approaches to observing the details of single-molecule interactions fall into categories based on the detection method (1). Many are fluorescence-based single-molecule optical biosensors (2⇓⇓–5), although other specialized physical techniques have been used, including electrochemical sensors (6, 7), plasmonic sensors (8), surface-enhanced Raman spectroscopy (9), and methods coupled to nanopore detection (3, 10, 11). In addition to complications of labeling procedures needed to add fluorescent reporters to targets of interest, single-molecule optical methods suffer from fundamental limitations in signal and resolution. A major challenge for single-molecule fluorescence methods is obtaining high signal-to-noise ratio, because the rate of photon production from single dye molecules is restricted by illumination intensity limits and photo-bleaching (12), constraining both short-time and long-time measurements. Molecular motion effects and diffraction also limit the ultimate spatial resolution or density of multiplex optical reporters.
Moving away from photon-based detection to all-electronic detection can remove these fundamental constraints on signal-to-noise ratio, scaling, and bandwidth, and moreover is maximally compatible with implementation on modern semiconductor chip devices. It would be advantageous to measure molecular interactions on a complementary metal-oxide semiconductor (CMOS) chip to leverage their low-cost mass manufacturing, speed, and miniaturization. These are hallmarks of modern CMOS chip-based devices, such as portable computers and cell phones. Such on-chip devices also enjoy a durable roadmap for future improvements provided by 50 years of Moore’s Law scaling of CMOS chips, and corresponding chip foundry infrastructure and supply chains.
However, the full potential of this vision of moving molecular biosensing “on-chip” can only be realized by using a suitably compatible sensor concept. To this end, there are three fundamental sensor design principles to consider: manufacturability, scalability, and universality (see SI Appendix). The field of molecular electronics provides a conceptual solution to all of these challenges, wherein a single-molecule in a circuit would provide the fully scaled sensor to solve the More-than-Moore scaling problem common to sensor devices. Scientific advances on the electrical properties of molecules, as well as bioelectronic inspirations, led to the proposal in the early 1970s that single molecules could be engineered for use as circuit elements (13), to perform circuit functions such as a rectifier or switch. Due to limitations of nanofabrication technology, it was not until the late 1990s that the first single-molecule circuits were demonstrated experimentally (14). Interest in this field of molecular electronics expanded dramatically after that point (15⇓⇓–18), and it was proclaimed the scientific breakthrough of the year by Science in 2000 (19). There it was noted that integrating molecules into chips would be the critical advance needed for this new field to have broad impact.
The experimental study of single-molecule electronic sensing was initially based on carbon nanotube (CNT) sensor devices. Their potential as sensors for single-molecule interactions became apparent (20) initially in the context of sensing gas molecules (21) and then chemical reactions (22). Nuckolls, Shepard, and colleagues (23⇓–25) introduced a single-molecule sensor for DNA–DNA binding (hybridization) processes, by functionalizing a CNT with a single DNA oligomer “probe” molecule. Collins, Weiss, and colleagues (26⇓–28) showed that a CNT can be used for real-time monitoring of the activity of a single enzyme molecule attached to nanotube, including DNA polymerase enzymes. Unfortunately, at present there is no way to mass manufacture CNTs having precise structure and functionalizations, and despite decades of attention (20), there is also no established path to integrating them into manufacturable CMOS chip devices (29, 30). Thus, while CNT molecular wire sensors enabled pioneering work on single-molecule sensing, they do not satisfy the design principles for a CMOS chip sensor platform.
In contrast, an ideal molecular wire should allow precision engineering to provide a site-specific conjugation moiety for attachment of probe molecules, as well as to provide suitable end groups for self-assembly into the nanoelectrodes on a CMOS chip, and should be readily available through existing manufacturing processes. This limits the candidates to peptides, proteins, or DNA as molecular wires, as these are in fact the only conducting polymers for which there are well-developed precision synthesis capabilities, including extensive means of precision functionalization. Double-stranded DNA (dsDNA) helices (31⇓⇓⇓–35) and protein α-helices (36⇓⇓⇓⇓⇓⇓⇓⇓–45) have both been studied as molecular wires. Examples from direct current measurements through various (short) α-helixes in the literature (42) suggest a (long) 25-nm α-helix could exhibit currents in the broad range of 3 picoamp (pA) to 120 pA at 1 volt bias, depending on the amino acid sequence, buffer conditions, and the nature of the peptide–metal attachment. Detailed tunneling probe methods have also recently been used to study conduction through larger proteins (46⇓⇓–49). While not nearly as conductive as CNTs, these biopolymers have the great advantage for present purposes of allowing precision engineering using existing manufacturing capacity.
Results
The chip presented here is a CMOS integrated circuit chip that supports a scalable array of nanoelectrode-based molecular electronics single-molecule sensors, as illustrated in Figs. 1 and SI Appendix, Fig. S1. These sensors are based on a molecular wire “bridge,” spanning a ~20 nm gap between the tips of nanoelectrodes, which themselves connect into a current monitoring circuit for real-time measurement of the current versus time flowing through the bridge. For a specific sensor application, the bridge molecule is conjugated in a site-specific manner with a biomolecule of interest, which acts as a probe for its molecular interactions with a target molecule. The observed current pulses represent the electrical resistance changes induced by these dynamic interactions. Since these sensors are isolated single molecules, they reveal binding events discretely and with high sensitivity, without the need for labels. The result is a direct electrical measurement of molecular interactions with corresponding kinetics.
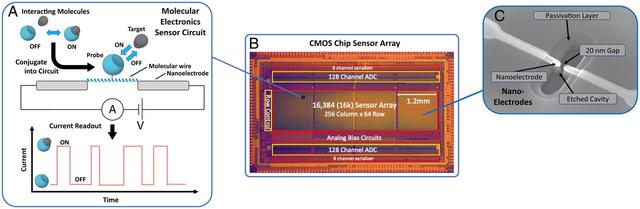
Fig. 1.Molecular electronic sensor and chip concept. (A) Senor concept: Given a pair of molecules that undergo an interaction, one of the pair is selected as a probe molecule and conjugated to a precision molecular wire (here a synthetic α-helical protein) that spans a nanoscale gap between metal nanoelectrodes. These connect it to a driving voltage source and current monitoring circuit to provide real-time readout of current vs. time, for the current passing through the molecular wire/probe complex. When the target molecule binds to the probe, the resistance of the complex changes, resulting in an observed change in current. The resulting current trace has on/off pulses that provide a direct representation of the molecular interactions. (B) CMOS chip device: A large-scale array of sensors are fabricated on the surface of a CMOS chip. Shown is an annotated image of the CMOS chip device used in these studies. This chip has 16,000 sensors and the circuitry needed to digitize and transfer sensor readings off-chip, at a rate of 1,000 frames per second. (C) SEM image of sensor nanoelectrodes, showing the 20 nm gap for the molecular bridge. Nanoelectrodes shown are fabricated by photolithography, using CMOS foundry-compatible process.
Fig. 2.The DNA hybridization binding sensor. (A) A single-stranded DNA (ssDNA) oligo probe, here the17-mer oligonucleotide (5′-TACGTGCAGGTGACAGG-3′), is conjugated to the bridge using conventional click chemistry at the 5′ end. (B) Example current vs. time trace, showing 6 s of data sampled at 1 kHz, taken with the sensor exposed to a 20 nanomolar (nM) concentration of the target oligo, here the complementary 14-mer strand (5′-CCTGTCACCTGCAC), suspended in a standard buffer solution. Each pulse of current above the baseline represents a single DNA binding event. The durations of the events and time between events are stochastic, with exponential distributions as summarized in SI Appendix, Figs. S2–S4. (C) The distribution of measured current values in the trace, shown alongside the trace as a vertical histogram, provides a visualization of the time spent bound (higher current) and unbound (lower current). In this example, the fraction of the time spent bound is 22%. (D) Response of DNA binding sensor to target concentration: binding of the 17-mer ssDNA probe on the sensor to the 14-mer target, at target concentrations of 10 nM, 100 nM, and 1,000 nM. The width of the peaks (dwell time) remains constant (~25 ms), but the time between peaks decreases from 45 to 4 ms as the concentration is increased, reflecting more frequent concentration-driven interactions. The fraction of time in the bound state (labeled “fraction bound” in plots) was estimated from the current measurement value histograms (Insets at Right). (E) Measurements of dwell time and fraction bound vs. concentration. As expected for DNA hybridization binding, dwell time remains constant with concentration, but fraction of time bound shows a classic saturation curve, from which is computed a binding affinity, Kd, of 39 nM. (F) Single-molecule thermal melting curves derived from the DNA binding sensor. In this experiment, a 45-mer ssDNA probe (5′-CGATCAGGCCTTCACAGAGGAAGTATCCTGTCGTTTAGCATACCC-3′) is attached to the bridge at the 5′ end. Two different complementary target oligos that are closely matched in properties but having different melting points were constructed by using a 15-mer target (5′-CCTCTGTGAAGGCCT) of the 45-mer, and an extension of this to a 20-mer target (5′-CCTCTGTGAAGGCCTGATCG). These were added to the chip at a concentration of 20 nM, performed in series. For each solution, the solution temperature was swept, in 2 °C steps, from 41 °C to 55 °C. Standard DNA melting curves were fit and used to derive the empirical melting points, Tmobs, shown. The results agree with the classical bulk predictions for the difference in Tm between the oligos, but here are measured entirely in a single-molecule context. (G) Specificity of the DNA binding sensor for mismatched DNA. The sensor was used to probe mismatched targets, using the 45-mer hybridization probe. The targets were 20-mers having 0, 1, 2, or 3 mismatches as shown (sequence Inset), added sequentially. The result shows a significant downward trend in fraction bound, as the number of mismatched nucleotides is increased.
Each sensor circuit (or “pixel”) is a dedicated current meter, implemented as a CMOS transimpedance amplifier that amplifies picoamp (pA) scale currents to millivolt (mV) scale voltages. The amplifier gain is 5 GΩ, so that each picoamp of current through the molecule is amplified to 5 mV of output voltage. The amplifier circuit in effect measures the current flowing through the biomolecule. The dynamic range of measurement is from 0 to 400 pA, and the amplifier is designed to be highly linear over this range, with subpicoamp noise (SI Appendix, Fig. S1 A and B), to preserve the detailed shapes of the sensor pulses for subsequent analysis. The sensor circuits are arrayed on the chip with a 20-μm pitch, for a total array of 16,384 (16k) sensors in an area that is just 1.2 mm by 4.8 mm (Fig. 1). The chip transfers the analog measured currents from the sensor array to on-chip analog-to-digital converters for digital transfer off chip, at a rate of 1,000 frames per second, and with 11 bits of digital precision for each current reading. The present chip is fabricated at a CMOS foundry employing a 180 nm “node” manufacturing process (the node is loosely related to the minimum transistor feature size on the chip, and more directly related to the relative density of circuits). As shown in SI Appendix, Fig. S1 A–D, the nanoelectrodes are fabricated in a second processing phase, using CMOS foundry-compatible photolithography processes and tools, to enable full integration of manufacturing into existing commercial CMOS chip foundries. Further details on the chip are provided in (SI Appendix, Fig. S1 C–F).
To illustrate the breadth of different types of molecular interactions that can be detected on the platform, this report demonstrates probes that detect binding of DNA (Fig. 2A), as well as protein molecules, small molecules, aptamers, and antibodies (Fig. 3 A, C, E, and G), and probes to monitor the activity of enzymes that are important for both applications and fundamental biology, e.g. CRISPR/Cas enzymes (Fig. 3I) and DNA polymerase (SI Appendix, Fig. S6A). SI Appendix, Fig. S7 A–F shows detailed ribbon models of all these sensor constructs. Direct imaging of several bridge molecules is also shown in SI Appendix, Fig. S7G.
Fig. 3.A survey of diverse molecular electronic sensors for binding and enzyme activity, shown to scale in molecular renderings, along with corresponding summary experimental results. (A–D) Protein and small-molecule binding kinetics. As a model system for showing protein binding and small-molecule binding, this sensor is configured to observe (A) a DNA polymerase binding a primer/template, and (C) a nucleotide binding into the polymerase pocket. For this model system, a 17-mer ssDNA template is conjugated to the peptide bridge at its 5′ end (and with 3′ end blocked to prevent the polymerase from binding that site). A complementary 14-mer primer strand is then bound to this, on the distal end of the 17-mer, to create a primer site on the sensor with the 3′-OH available for polymerase binding. (B) Summary kinetics (dwell time, fraction of time bound) for Klenow DNA polymerase binding to the primer site, as polymerase concentration is titrated from 0.008 to 3.8 µM, in a background of 100 nM 14-mer primer to suppress primer dissociation. The inferred binding affinity of the polymerase is Kd = 530 nM. (D) A nucleotide titration is performed to observe the binding in the polymerase pocket, in a noncatalytic buffer so as to observe the binding kinetics without incorporation. A 45-mer template is on the bridge, and a 31-mer primer was bound to the distal end of the template, so that the first template base (A) is complementary to the nucleotide being tested (T). The nucleotide was added in concentrations of 2.5, 5, and 15 µM along with the 100 nM polymerase and primer in the presence of a buffer that has 10 mM Sr2+ (without Mg2+), in which nucleotide incorporation cannot occur. In this buffer, the dNTP will repeatedly bind and dissociate from the polymerase pocket, and the resulting summary binding kinetics are shown. This also serves to illustrate the detection of small molecule binding. (E and F) Aptamer sensors: Aptamer sensors were constructed, here targeting the SARS-CoV-2 S protein with a 57-mer DNA aptamer (E) and targeting the SARS-CoV-2 N protein with a 97-mer DNA aptamer (SI Appendix, Fig. S7B-2), both taken from the literature. (F) The concentration response titration curves for both the S aptamer and N aptamer sensors, for a range of applied target protein concentrations. The binding affinities, Kd, derived from these curves (6.4 nM, 39 nM) are similar to those reported for standard bulk aptamer binding assays in solution. (G and H) Antibody–antigen sensors. As a model system, a fluorescein–antifluorescein antigen–antibody pair was used, with the fluorescein antigen presented on the sensor by tethering it to the bridge using a ssDNA oligo as a linker. A 45-mer oligonucleotide was used, with the 3′ (distal) end of the DNA capped with a fluorescein during synthesis. A commercial antifluorescein antibody (Fab) was added in TKS buffer on the chip. The summary kinetics are shown for dwell time and fraction of time bound, as the concentration of antibody is titrated over the range shown. The inferred binding affinity was Kd = 1.3 µM. It was observed that all binding signals were extinguished when 4 mM of free fluorescein was added to saturate the antibody, verifying the specificity of the binding signal. (I and J) A CRISPR/Cas enzyme activity sensor. To assemble a Cas enzyme as a probe on the bridge, first a guide RNA targeting a dsDNA target for a CRISPR/Cas12a enzyme was conjugated to the bridge, and these were assembled on chip. A Cas12a enzyme was provided in solution and allowed to dock to the guide RNAs on the bridges, thereby programming it for the target dsDNA, and also effectively tethering it to the bridge as a probe. For these experiments, the guide RNA is a 40-mer, attached to the bridge peptide using click chemistry at the 13th nucleotide, which is the base that extends furthest outside the enzyme in the pseudoknot loop. The kinetics are summarized in the titration curve, showing fraction of time bound saturating as the dsDNA target varies in concentration, in the presence of a concentration of 20 nM free (untargeted) Cas12a enzyme. Thus, this configuration acts directly as a sensor for the dsDNA target, without assessing posttarget-binding nonspecific single-stranded nuclease activity. This latter nonspecific activity is also observable on the sensor when provided with a ssDNA substrate. The observed binding affinity for the dsDNA target is Kd = 3 pM. The experimental buffer was 20 mM Tris·HCl pH 8.0, 20 mM KCl, 10 mM SrCl2, 4 mM DTT. Two examples of raw sirgnal traces for D (small molecule) and E (protein) are included in SI Appendix, Fig. S7 B and F, showing that the character of the bridge current signals is similar for these diverse probes.
All these sensors have a common format, consisting of a probe molecule precisely conjugated to a molecular wire (Fig. 2A). The molecular wire bridge used in this work is an α-helical peptide, 25 nm in length, with a specific conjugation site engineered into the side chain of an amino acid near the middle of the peptide for attachment of probe molecules, and metal-specific conjugation groups (1) engineered on to the ends for selective binding and self-assembly to the metal nanoelectrodes. When the bridge molecule is delivered to the nanoelectrodes by passive diffusion, such assembly can take ~24 h (e.g., using 20 nM bridge concentration and dilute buffer [2 mM Tris pH 7.5]). In contrast, for all the work presented here, an “active bridging” dielectrophoresis protocol is used to greatly accelerate and enhance the assembly process. Dielectrophoresis is a highly efficient process in which electrical forces attract polarizable molecules to the gap between micro- or nanoelectrodes (31, 35, 36, 50, 51).
In the present work, this allows assembly to be completed in under 10 s. The dielectrophoretic trapping protocol relies on the application of an AC voltage, as a 100 kHz, 1.6 V peak-to-peak sine wave, applied for 10 s, followed by a DC current measurement to assess bridge formation. Up to 10 such cycles are applied in the bridging process. These trapping voltages are simultaneously applied to all nanoelectrodes on the chip, by switching in on-chip AC driving circuits. Compared to passive diffusion, dielectrophoretic trapping shortens bridging time to ~10 s (~10,000-fold faster), while simultaneously working at ~1,000-fold lower input concentrations of bridge molecules. This enhancement in the reaction rate suggests the trapping field effectively increases the local concentration of the bridges near the electrode gaps by at least 1 million-fold. To assess effectiveness of bridging, the sensor DC current after bridging is compared with the value prior, and a sufficient jump in current is indicative of successful bridging. A population of sensors showing substantial bridge current increase is thereby observed, typically over 10% of all available pixels on the chip, indicating the presence of the 25 nm peptide bridge spanning the electrode gap.
DNA Oligo Hybridization Binding and Sensor Validation.
The sensor and chip performance are validated here using the well-studied model system of a single-stranded DNA oligo as a probe for hybridization to its complementary strand. This is a useful reference system, since oligo binding has been extensively studied, both empirically and theoretically, including at the single-molecule level using CNT sensors (23⇓–25).
For the probe, a 17-mer DNA oligonucleotide was attached in a precision site-specific manner to the central amino acid on the bridge (Fig. 2A), using conventional click chemistry (Materials and Methods). Once the bridge molecules (with probe) are attached to the electrodes of the chip, baseline current is measured, which is typically several picoamps under applied voltages in the range of 700 to 1,000 mV. While continuing to monitor sensor currents, the “target” complementary 14-mer oligonucleotide is added at a particular concentration. As shown in Fig. 2B, the sensor on chip responds to the presence of the target in solution with current pulses that can be interpreted as individual binding events between the conjugated probe ssDNA and its complementary target strand present in solution. These exist in a dynamic equilibrium between bound and unbound states. Control experiments reveal that this binding changes with the target concentration (Fig. 2D) and temperature (Fig. 2F), as would be expected for DNA–DNA hybridization.
Analysis of single-molecule binding data using hidden Markov models.
Hidden Markov Models (HMM) are used to quantify the primary signal traces. HMM have previously been successfully applied to timeseries data from single-molecule biophysics experiments (52⇓⇓⇓–56). In the present case, the HMM assigns the “hidden” bound and unbound states of the sensor to segments of the observed signal trace that have statistically different current levels, typically with lower currents for the unbound state and higher currents for the bound state. (See SI Appendix, Materials and Methods for HMM details and kinetic parameter estimation.) The HMM segmented signal trace allows extraction of the individual waiting times between binding events, τ0, and the individual dwell times or time spent bound, τ1 (SI Appendix, Figs. S2 and S3). These times obey exponential distributions (SI Appendix, Figs. S2 and S4), as expected for two-state first-order chemical reaction kinetics. A convenient summary statistic is the total fraction of time spent in the bound state (denoted “fraction bound” in all figures; e.g., Fig. 3 B, D, F, H, and J). This is seen to scale with concentration according to a classic Michaelis–Menten saturation curve (Materials and Methods). Fitting this curve allows calculation of the classic binding affinity of the interaction, Kd, which at the single-molecule level can be defined as the target concentration at which the single probe molecule spends equal time bound and unbound. The fraction of time spent in the bound and unbound states can also be conveniently visualized using vertical histograms of all the measured current values in a signal trace segment, as shown to the right of the traces in Fig. 2 C and D.
Single-molecule thermodynamics: melting curves.
Another application of the sensor is to determine the melting temperature, Tm, of the DNA duplex, which is defined at the single-molecule level as the temperature at which the probe DNA molecule spends equal amounts of time in the bound and unbound states, for a given concentration of target oligo. As shown in Fig. 2F, measuring fraction of time bound at eight temperatures allows fitting of the data to a classic DNA hybridization melting curve. For these experiments, a Peltier heating plate in direct thermal contact with the chip is used to set different temperatures in succession, with the measured temperatures for the chip provided by on-board temperature sensors. From these curves, it is clear that a 20-mer target oligo melts at a higher temperature than a 15-mer subsegment, as expected. Determination of Tm serves to validate that the sensor measures the DNA–DNA hybridization binding reaction as intended, and also has practical value in selecting a suitable operating temperature for making measurements of many binding events. As is done classically, this single-molecule melting curve can be used as a measurement modality to enhance detection specificity for the target of interest, or to characterize targets that contain mismatches.
Mismatch sensitivity.
The single-molecule binding probe signal trace contains rich information about the binding reaction and is also highly sensitive to the specific binding target. This can be illustrated in fine detail for DNA oligo binding by looking at the impact of single-base mismatches introduced in the target oligo sequence. As shown in Fig. 2G, four variants of a 20-mer target DNA were made, having from 0 to 3 mismatched bases. With each additional mismatch, the fraction bound decreases (and off-rate koff increases). These differences in binding kinetics could be further magnified by performing the measurements at a temperature nearer to the Tm of the matched target, or by performing a temperature melting curve, as in Fig. 2F. This sensitivity to mismatches can have applications for DNA binding assays in which sequence variants relative to a reference sequence probe might be of interest, such as in detecting novel strains of a viral genome, detecting somatic mutations in a cancer genome, or detecting SNP genotype variants.
Sensor signal generation mechanism.
CNT biophysics experiments with a DNA probe (23) as well as enzyme probes (26, 57) show that the dominant signal generation mechanism in single-walled CNT sensors is a field-effect, where electrical fields emanating from the target-probe complex alter the flow of current in the CNT wire in an action-at-a-distance manner. This is demonstrated by increasing salt concentration to screen out electric fields in the solution, and showing the signal is reduced from charge groups more distant than the screening (Debye) length (23). For the present molecular electronic sensor, similar experiments were run to examine the effect of increasing salt concentration in the binding buffer on both signal strength and dwell time, with salt concentration (KCl) increasing from 2 mM to 2,000 mM (Debye length decreasing from 6.8 nm to 0.22 nm). As shown in SI Appendix, Fig. S5, the signal strengths (defined here as pulse heights) decrease by nearly 70% with salt concentration. This suggests that the dominant part of the current modulations observed are due to the field effect demonstrated for CNT sensors. There may be other minor forms of conduction modulation also involved. For example, these may relate to direct changes in electron conduction paths through the molecular complex, or to changes in the ion clouds in the solution around the molecules, or to electrochemical state changes on the molecular sensor, such as redox charge transfer reactions. In any case, one benefit is that the sensor allows for measurements of binding even in biologically relevant high salt conditions, much higher than is possible for CNT sensors (23).
Protein and Small-Molecule Binding.
Fig. 3 A and C show two binding processes fundamentally related to DNA polymerase: the binding of the protein to a 3′-OH primer site (Fig. 3 A and B), and the nucleotide substrate binding in the active pocket of the enzyme (Fig. 3 C and D). These particular sensor modalities are useful for the study of polymerases, but they also serve to illustrate detection of protein binding (here polymerase docking to a priming site) and small-molecule binding (here a dNTP interacting with the polymerase binding pocket). The latter could be considered as a model for a small-molecule drug interacting with a binding pocket on a protein target. To construct this sensor, a DNA template oligo is first tethered to the bridge, as above in the DNA binding studies. A complementary primer oligo is then bound to this, to form a primer site, which can then act as a probe for binding a polymerase. Titration of Klenow DNA polymerase binding to the primer complex on the sensor produces a typical saturation curve (Fig. 3B), and a resulting binding constant, Kd. The binding of a nucleotide substrate to the binary complex of primer-polymerase is measured (Fig. 3D) in a noncatalytic buffer (lacking Mg2+), so that the polymerase cannot incorporate the nucleotide. Under these conditions, the sensor measures the dynamic equilibrium of the nucleotide substrate entering and exiting the binding pocket, producing a corresponding binding signal trace, and titration saturation curve (Fig. 3D).
Aptamer–Target Binding.
A DNA aptamer is an oligomer with a sequence empirically selected to bind a specific protein or molecule of interest. Such aptamers can be attached to the sensor bridge as probes in exactly the same manner as the oligonucleotides used for DNA binding probes (Fig. 3E). The use of aptamer binding probes provides the capability to detect a great diversity of possible targets. In particular, aptamer technology is well-suited for rapidly developing binding probes for protein targets, for use in targeted protein detection and identification, and proteome characterization and profiling.
One particularly timely application of such protein-binding aptamers is in testing for infectious disease pathogens (58). In particular, the COVID-19 pandemic has highlighted the need for cost-effective, highly specific, rapid, and distributed testing for viral particles. Aptamers against the SARS-CoV-2 N gene protein (nucleocapsid phosphoprotein) (59) and the S gene protein (surface glycoprotein or Spike protein) (60) have been described in the literature, which exhibit the selective and high-affinity binding necessary for a diagnostic test. Antigen tests have been developed using these aptamers, which show high accuracy in detecting SARS-CoV-2 infection (61). To test the use of aptamers on the present sensor chip platform, both the N and S protein aptamers were attached to the bridge (Fig. 3E and SI Appendix, Fig. S7 B and B-2) to study their binding kinetics against SARS-CoV-2 proteins. Using aptamer-functionalized bridges on chips, the corresponding N or S protein target concentration was titrated in solution (Fig. 3F). The measured binding kinetics parameters from chip experiments for the N protein were koff = 40 s−1, kon = 6.2 × 108 M−1 s−1, and Kd = 64 nM. Similarly, measured parameters for the S protein were koff = 35 s−1, kon = 3.3 × 109 M−1 s−1, and Kd = 6.1 nM. These data are in good agreement with the reported affinities for the N protein and S protein aptamers, which were 20–200 nM and 5.8 nM, respectively.
Antibody–Antigen Binding.
To demonstrate antibody–antigen binding, one option is to place the antigen on the bridge and observe the binding to the cognate antibody present in solution. A model system for this is to use a fluorescein dye molecule as the antigen, and a commercially available antifluorescein antibody, as illustrated in Fig. 3G. The fluorescein is conveniently mounted on the bridge using a DNA oligo as a linker, attached by the same chemistry as described above, and where the DNA oligonucleotide is synthesized to have the fluorescein molecule at its 3′ (distal) end to present the antigen. A commercially available antifluorescein antibody was titrated on this chip and the binding activity is summarized in Fig. 3H, resulting in an apparent Kd of 1.3 µM. A DNA oligo tether, such as used here, is one convenient means of conjugating antigen molecules to the bridge; however, many well-known methods of conjugation can be incorporated into the bridge molecule design, to support attaching antibodies or antigens, in particular conjugation methods that are compatible with existing antigen or antibody libraries.
CRISPR/Cas Enzyme Activity.
The CRISPR/Cas enzymes originally used for gene editing (62, 63) have recently been proposed as tools for sensitive DNA detection for diagnostics and other applications (64⇓⇓–67). In general, a CRISPR/Cas enzyme binds to a short-guide RNA strand that serves to program it for sequence-specific binding and activation, for example, to cleave the specified target DNA strand. The ability to monitor the single-molecule kinetics of these enzymes could be useful to understanding their multiple complex activities, and also may help in enzyme evolution studies to provide high-throughput screening for useful mutant phenotypes (68⇓–70). In addition, such enzymes have potential uses for diagnostics, based on monitoring for indications that the programmed enzyme has bound its specific target. The originally discovered Cas9 enzyme has been widely adopted for gene-editing functions, but more recently discovered Cas enzyme families—such as Cas12, Cas13, and Cas14—have been proposed for diagnostics (64, 66, 67), since they undergo more dramatic and readily detectible transformations after encountering their target, and therefore simplify the proposed optical reporter methods. However, the single-molecule sensor presented here is capable of directly observing the primary DNA target capture, and thus any of these enzymes could potentially be used diagnostically, and in a highly multiplex target fashion, on these sensor array chips.
Shown in Fig. 3 I and J are results from binding experiments using the CRISPR/Cas12a enzyme (64, 71), which is commonly used as the basis for such diagnostics, programmed by a guide RNA designed to detect a 20-base DNA sequence taken from the S gene of the SARS-CoV-2 virus. The resulting guide RNA bridges were assembled onto the chip, which was then first used to observe titration of the Cas12a enzyme binding to the guide RNA, over a protein concentration range of 0 to 1 µM. Binding was observed when the guide RNA was attached to the bridge at its 13th nucleotide (which corresponds to the outermost exposed point in the pseudoknot loop) (SI Appendix, Fig. S7D) but was not observed when the attachment was at the 3′ terminus of the guide RNA. It is likely that this latter configuration results in steric hinderance to Cas binding, based on the known Cas12a-guide RNA structure (Fig. 3I and SI Appendix, Fig. S7D). The chip with such guide RNA bridges was incubated with 20 nM Cas12a to bind and program the enzyme, and then titrated with 0.1 to 64 pM concentrations of target dsDNA, done under non-catalytic buffer conditions in order to observe the target binding kinetics. A binding affinity of 3 pM was observed for dsDNA binding to the Cas12-guide RNA complex, similar to the results reported for such enzymes (64, 65, 71, 72). Extending the concept illustrated here, tethering distinct guide RNAs to the bridges of distinct sensors on the array provides a means to deploy multiplex targeted CRISPR/Cas enzymes on chip, and monitor their primary detection activity as single molecules, in parallel. This capability could provide for highly multiplexed CRISPR diagnostics, or for massively parallel phenotype screening for Cas enzyme evolution (68).
DNA Polymerase Enzyme Incorporation Activity.
In this case, a single Phi29 DNA polymerase (73) molecule is conjugated to the molecular bridge, using a site-specific conjugation method. The resulting sensor-chip device is provided with a primed DNA template and the required dNTPs for the polymerase to extend the primer on the template. This sensor configuration, illustrated in Fig. 4 and SI Appendix, Fig. S7E allows single-molecule observation of the polymerase activity as it binds and incorporates nucleotides in real time. Exemplary signal results are shown in Fig. 4 and in more detail in SI Appendix, Fig. S6, which show a 25 s portion of the sensor signal trace having ~40 pulses with dwell times longer than ~10 ms. Inspection of this signal trace suggests that each major pulse represents a nucleotide binding and incorporation event, consistent with the known behavior of DNA polymerase and with CNT observations of single-molecule polymerase activity (26⇓–28). For the DNA template chosen here, primer extension should result in 15 dCMP incorporations followed by 25 dAMP incorporations. Visual inspection(Fig. 4) suggests there are two distinctive segments of pulses distinguished by features such as the pulse height, pulse width, and waiting time between pulses. These data segments are therefore putative A-pulse series and C-pulse series, which highlight the potential to distinguish A and C incorporation events. For a more detailed quantitative analysis showing 95% distinguishability of these pulses, see SI Appendix, Fig. S6A and related Methods.
Fig. 4.DNA Polymerase Activity Sensor. A phi29 DNA polymerase is conjugated to the sensor bridge using the SpyTag-SpyCatcher conjugation scheme shown. The 25-second-long signal trace shows an isolated burst of sensor activity that occurs after adding a primed 40-mer template (sequence 5'-25T-15G-3') and corresponding (dCTP and dATP) nucleotides. The expectation is the polymerase would acquire a template and incorporate 15 C's followed by 25 A's. A series of ~40 discrete major pulses are seen, representing putative incorporation events. The signal trace has ~15 wide-spaced, narrower pulses on the left (green region), and ~25 closely spaced, broader pulses on the right (red region), suggesting these are the C and A events, respectively, and that therefore the C and A incorporations events can be distinguished by examining pulse features.
Shown in SI Appendix, Fig. S6 B and C (and SI Appendix, Table S3) are more extensive pulse discrimination analyses, wherein a total of seven feature metrics of each pulse are automatically extracted, and subsequently assessed in a principal component analysis (PCA). When the extracted pulses from the trace are displayed using the two dominant PCs (SI Appendix, Fig. S6B), they fall into two nearly disjoint clusters based on these shape features. These clusters also correspond to the pulses from the putative oligo-dC segment (SI Appendix, Fig. S6B, coded green) and oligo-dA segment (SI Appendix, Fig. S6B, coded red) of the signal trace. Thus, the pulses are largely distinct in their features, consistent with the hypothesis that dAMP and dCMP incorporations produce distinctive pulses. The PCA importance weighting chart (SI Appendix, Fig. S6C) shows the relative contribution of the various shape features to the first PC (SI Appendix, Table S3). This example illustrates the potential for sequencing DNA using such a DNA polymerase activity sensor, based on analyzing the detailed single-molecule kinetics and signal pulse features of the polymerase as it copies a template.
Sensing in Complex Backgrounds: A Model Viral Detection Assay.
The basic detection of molecular interactions demonstrated above can be used to develop many applications. One such example is viral detection, which has been highly relevant to the COVID-19 pandemic. However, for use in practical diagnostic tests, it is important that a sensor be able to reject complex backgrounds of off-target molecules. Ideally, the sensor would be able function in crude samples, such as saliva. To test this, we constructed a model assay mirroring the Center for Disease Control and Prevention’s (CDC) issued recommendation for a qPCR test for the COVID-19 infection, based on testing for the presence of two sequences in the N gene (74). PCR products were made using the CDC forward and reverse primers for SAR-CoV-2, with a synthetic plasmid for the N gene serving as the positive control target template, which was spiked into contrived samples. The sensor chip was prepared with a ssDNA oligo probe targeting one strand of the PCR product. This resulting PCR product, unpurified, was applied to the chip to assess the ability of the DNA hybridization sensor to reject complex backgrounds and still detect its specific target, as well as to work with the complex mixtures produced by PCR, as would arise in practical diagnostic tests (Fig. 5). The oligo probe on the bridge is a 21-mer (CCGCATTACGTTTGGTGGACC) taken from the CDC qPCR TaqMan probe sequence (2019-nCoV_N1-P: FAM-ACC CCG CAT TAC GTT TGG TGG ACC-BHQ1) (74). Various target sequences tested against this probe are shown in SI Appendix, Table S2.
Fig. 5.Using a DNA binding sensor to detect a viral target under mock assay conditions. The DNA binding probe chip is used to detect a target DNA PCR product produced from a contrived sample, with confounding complex background. (A) The titration curves show chip sensor response, for various concentrations of targets, for targets consisting of a synthetic 24-mer positive control, an unpurified PCR product from a contrived saliva sample, and this PCR product with salmon sperm DNA added at 2 μg/mL, to mimic the impact of having background genomic DNA contamination in saliva. This high, complex background had little impact on the results. (B) A 3 s signal trace at the lowest concentration tested, 100 pM, showing 5.2% fraction of time bound. This illustrates the strong signal spikes, and the potential to detect much lower concentrations through longer observations.
Each of these targets (SI Appendix, Table S2) were tested for binding on chip and found to conform to positive and negative controls, as expected. Only the last example (double stranded PCR product) displayed weaker binding than the others, suggesting it has partially renatured (thereby lowering the concentration of target single strands) prior to testing on chip.
Targets were tested at concentrations of 10 pM, 100 pM, 1 nM, and 10 nM in buffer A. The PCR products registered similar sensor responses to pure oligo target samples (Fig. 5A), even though they represent a much less pure sample because of PCR off-target byproducts and reagents. In order to further assess the impact of complex background materials, buffer A was mixed with heat-inactivated human saliva (from 10% by volume to 50% by volume), and in another trial it was mixed with a high concentration of highly complex background DNA: salmon sperm DNA at a concentration typical for DNA contamination of saliva (2 µg/mL) (Fig. 5A). Neither one of these challenges had substantial impact on the sensor readout, showing that the sensor is highly specific for its target and robust against complex and even crude saliva samples.
In terms of the overall sensitivity, inspection of the signal at the lowest tested concentration, 100 pM, shows (Fig. 5B) the signal consists of clear pulses of ~6 pA magnitude occurring approximately eight times per second, but bound just 5% of the total time. This target concentration corresponds to ~1 femtomole of oligonucleotide applied to the chip. Based on the trace shown, this is not the lower limit of on-chip detection: at even lower concentrations, by observing such a sensor for longer time, or observing multiple such sensors in parallel for the same target, it would be possible to observe a statistically meaningful number of binding event pulses at much lower target concentrations. This provides a powerful means to reduce the limits of detection for such a test, potentially by up to six orders of magnitude.
Discussion
The molecular electronics sensor chip presented here has a number of powerful features, as well as broad potential for future applications. In particular, as a sensor platform, it has a unique combination of broad applicability, scalability, and single-molecule sensitivity, while the CMOS chip format also provides for manufacturable realization of sensitive, multiplex, rapid, low-cost tests, on compact instruments. These combined features could enable attractive near-term applications, from drug discovery to diagnostics to DNA sequencing. Moreover, it may provide these with a long-term, faster-than-Moore’s Law scaling path to ever lower costs and greater speeds for highly durable technology solutions. These points are briefly discussed further in the following sections.
Sensitivity.
The molecular electronics sensor has intrinsic single-molecule detection sensitivity. This is a relatively unique capability in biosensing, where methods that have single-molecule detection capability often rely on biological signal amplification (such as in PCR or ELISA) to increase the signal to the point where it can be readily detected. Having the platform based on a true single-molecule sensor provides the potential for the ultimate limits of sensitive detection, with or possibly without combination with biological signal amplification for various assays. Moreover, while not tested here directly, it may be possible to further increase sensitivity by electronic target amplification using dielectric forces to concentrate targets, as was used here to increase bridge-electrode assembly efficiencies as much as a million-fold.
It is known that such highly sensitive electronic amplifier systems can experience random telegraph noise (RTN) resulting from, for example, single trapped charges changing state. Indeed, the signal pulse trains produced by the molecular electronics sensor (e.g., Fig. 2B) look similar to RTN. Such spurious RTN noise modes do indeed appear sporadically on the present type of sensors, and therefore it is important that experiments be designed with controls to distinguish these spurious noise modes from proper detection signals. One useful method is to titrate the concentration of the target and verify that the sensor pixel responds properly to this titration. For example, Fig. 2D shows the output for different target concentrations for the DNA hybridization sensor. This can thereby rule out pixels subject to RTN that could otherwise confound measurements. In addition, the dwell times for RTN pulses will not match the expected dwell time or temperature response for the target of interest, and this can further be used to reject RTN artifacts. Such calibrations and quality controls should ideally be built into assay protocols.
Specificity.
The molecular electronic sensors can provide highly specific detection, as long as the primary molecular interaction is specific. This is perhaps surprising, given the extreme sensitivity of the sensors. This specificity is best demonstrated with the DNA hybridization binding sensors, where the strength of the DNA binding reaction is in effect sequence-programmable, and finely tuned off-target interactions can be studied by introducing mismatched bases in the target. The sensors were challenged with such complex interactions to demonstrate the limits of specificity. As shown in Fig. 2G, a matching oligonucleotide target produces a signal trace distinguishable from one possessing even a single mismatching nucleotide. The difference in affinity is readily apparent by observing the fraction of time bound and also dwell time. This also has implications for practical assays, since such binding probes can therefore be used to discern the presence of a SNP for genotyping, or to detect isolated mutations in viral genomes for viral strain detection, or for cancer genome mutation analysis. At the other extreme of specificity, the sensor demonstrated the ability of the probe to reject highly complex off-target backgrounds, such as genomic DNA and crude saliva. This has implications for practical assays, enabling them to be robust and have simple, rapid sample preparation.
Multiplex Measurements.
The generality of the sensor platform was demonstrated by measuring enzyme activity (DNA polymerase and Cas12a nuclease), and binding of aptamers (for SARS-CoV-2 S and N proteins), an antigen (fluorescein) with antibody target, as well as binding of proteins and small molecules. The scalable pixel array chip therefore brings powerful and practically unlimited multiplexing capabilities to all of these types of detection. For example, it was shown that a DNA binding probe could detect a gene from SARS-CoV-2; through such multiplexing, similar DNA binding probes could target many different viruses or viral strains on one chip, applied to a single sample. Since each pixel is an independent sensor, even the present chip provides a capacity to multiplex from 1 up to 16,000 probes, and future scalability can readily take this to millions and well beyond (see Scalability below). Methods for constructing such multiplex probe arrays include classic methods, such as fluidic partitioning, or random deposition and combinatorical decoding, such as are used in DNA microarray technology. However, the electronic sensor pixels provide an option of voltage-directed assembly of different probes to specified pixels, by only activating the trapping voltage at pixels of interest as each different probe solution is serially introduced to the array. This would enable precision multiplexing of up to hundreds of probes, due to the speed of active trapping.
Utilizing the full pixel capacity of such chips for massive multiplexing is important for applications such as whole-genome DNA sequencing or whole-proteome profiling, where the anonymous single-molecule targets are captured at each sensor site (DNA target or protein target, respectively) and there undergo many interrogation reactions (polymerase processing, or antibody/aptamer response profiling, respectively) to fully identify or characterize each anonymous DNA or protein target. These extremely high-throughput applications can directly benefit from the multiplexing provided by future chips that scale to millions or even billions of sensors.
Scalability.
The present sensors solve the More-than-Moore scaling problem: the physical extent of the sensors here is defined by the bridge, which is 25 nm long (and which was chosen to be large enough to accommodate all biomolecules of interest as probes). This is already substantially smaller than the minimum metal pitch (MMP, the closest possible metal electrode spacing at the first contact layer used to make contact to transistors) on all existing CMOS fabrication nodes (MPP is ~36 nm on the state-of-the-art [circa 2021] 5 nm CMOS nodes), as well as for all projected nodes spanning the next 10 years (3 nm, 2 nm, 1.4 nm) (75, 76), and thus the size of the molecular element in no way limits the ability to shrink the CMOS pixel circuits (from the current 20 μm pitch reported here, fabricated on a 180 nm CMOS node) in current or foreseeable foundries. Indeed, allowing for reasonable engineering feasibility in existing foundries, future molecular electronic sensor pixels could have a limiting pitch approaching 100 nm, which still allows ample room for the nanoelectrodes contacting vias, and a number of transistors for circuit implementation. This corresponds to a density of 100 million sensors per square millimeter of CMOS chip die. Thus, chips the size of a square millimeter, costing pennies, could provide fantastic sensing capacity. Conversely, a large chip, such as a 10 mm × 10 mm sensor array die, would provide for up to 10 billion sensors.
Rapid Detection.
The single-molecule sensors demonstrated here expose the dynamic nature of single-molecule binding interactions. Just a few seconds of data can survey enough binding events to gather quantitative statistics (e.g., Fig. 2B), which can enable extremely rapid measurements and rapid testing (77). In general, near the chemical equilibrium point of the interaction (e.g., Tm for DNA–DNA binding), the rate of binding nearly equals the rate of unbinding, and numerous events can be observed in a short time, such as the span of seconds. Controlling key reaction variables, such as temperature and target concentration, can be used to adjust the interaction kinetics into such a desirable regime for rapid testing.
Low-Cost Tests.
In many use cases, such as for diagnostics, the CMOS sensor chip would be a single-use disposable. Because of the economy of scale of manufacturing, CMOS chips are extremely low cost when produced at high volume, and therefore support low-cost testing. For example, circa 2021, in the 180 nm CMOS node foundries, medium-to-high production volume, finished commercial CMOS 200 mm wafers cost in the range of $1,000 to $1,400 per wafer, or ~4 cents per square millimeter. (For example, the chip die in the present studies is ~25 mm2.) Finer fabrication nodes—such as 65 nm, 22 nm, and 7 nm—are only several-fold more expensive, while enabling orders-of-magnitude higher sensor densities. As shown in Fig. 1B, each square millimeter can contain thousands—and potentially many millions—of sensors (see Scalability above). Thus, molecular electronics chips enable extremely low-cost diagnostic tests.
There have been recent proposals for penny-scale diagnostics relying on low-cost materials, such as PDMS, paper, and ink-jet printed nanoparticles (78⇓–80), yet the extreme economics of CMOS manufacturing allow millions of highly sophisticated sensor circuits to be fabricated on pennies-worth of finished CMOS. The potential of CMOS to provide extremely low-cost diagnostics should not be ignored when paired with general and maximally scalable electronic sensors. In addition to low-cost production, it is important to note that production capacity is unparalleled: the global foundry capacity is estimated to be the equivalent of several hundred million wafers per year, and the industry currently delivers over 1 trillion chips per year. Even for the most extreme imaginable high-volume testing scenarios—such as a future pandemic where such chips are used to test the global population on a near-daily basis—the CMOS chip industry, uniquely, has the required manufacturing capacity. Even for diagnostic concepts based on simple, low-cost materials, in the absence of a manufacturing base it can take decades to reach these production scales.
The instrument needed to run CMOS chip-based assays can be compact, comparable in size to a portable computer or cell phone or USB stick, so that diagnostic tests could be run at the site of use, such as medical point-of-care, or transportation hubs or other public sites, or in homes. For reference, the instrument used for the experimental work reported here (SI Appendix, Fig. S1C) is smaller than a laptop and yet was not optimized for small size. This potential for ultrasmall form factors could support novel environmental sensing methods, such as drone-deployed pathogen sensors actively surveying air or wastewater. Compact, low-cost electronic devices, comparable to a digital thermometer, could also be suitable for home-screening assays of general interest, like health and wellness biomarker panels, early indicators of disease, and at-home diagnostics.
Applications.
These general features of the platform provide support for many applications, with the potential for attractive capabilities and dramatic performance improvements. A few notable examples are discussed here to illustrate this potential.
Molecular electronic microarrays.
The hybridization sensor presented here, deployed in a massively multiplex fashion with many different hybridization oligos represented on one sensor array chip, provides the molecular electronics equivalent of a classic DNA microarray, and could be used for many of the same applications (81⇓–83). However, recast in this framework, it also provides the features of rapid readout and real-time, label-free detection, in an all-electronic format compatible with field deployment on compact devices. Such a next-generation microarray thus confers many benefits. If the oligo probes are taken to be aptamers, this can further provide for diverse targeting, such as protein detection arrays. This illustrates the general principle that mature, classic binding assays, when their molecules are recast as molecular electronic sensors on such a chip platform, inherit many major performance advantages, as well as a long roadmap of further performance improvements.
Drug-target interaction characterization.
The ability to do label-free, time-resolved detection of small-molecule–protein and antibody–antigen interactions enables drug-discovery applications (84). In particular, this may be especially useful for characterizing very weak binding interactions that may represent the earliest stages of drug-candidate selection for poorly druggable targets (85⇓–87). In addition, since the chips are sensitive to single molecules, assays may operate with minimal input materials to support testing of rare compounds. The potential for massive multiplexing on-chip could translate into efficient high-throughput screening of drug candidates, or for molecular evolution programs that rely on screening many mutant protein phenotypes, such as for antibody engineering, developing new CRISPR/Cas genome-editing enzymes, or directed evolution of proteins (68⇓–70, 88).
Diagnostic testing.
The basic sensor types demonstrated here provide a unifying foundation for transferring content from existing molecular diagnostics platforms (77). For example, DNA hybridization is the basis of many forms of nucleic acid detection, such as in qPCR or DNA microarrays, as used in nucleic acid tests for viruses and infectious disease pathogens. Antigen–antibody binding (or aptamer binding) is the basis for immunoassays, as commonly used in lateral flow detection of various antigens, such as pregnancy tests, detection of protein biomarkers, or screening of panels of molecular allergens in the diagnosis of allergy and autoimmune disorders. CRISPR/Cas enzymes have recently been proposed as the basis for new types of diagnostics (64, 66, 67). The sensor examples shown here offer the potential to unify all these disparate diagnostics onto a common chip platform, and provide the benefits of highly multiplex, low-cost tests on a deployment format well-suited to point-of-care testing. Included in this unification is DNA sequencing, which is a diagnostic modality of special importance due to its fundamental role in precision medicine (89).
DNA sequencing.
The binding sensor with a DNA polymerase probe ( Fig. 4 and SI Appendix, Fig. S7) can monitor the activity of the polymerase as it copies a template, with resolution of the individual nucleotide addition events, and discrimination between bases. The ability to monitor a polymerase generally enables “sequencing by synthesis” methods, such as first introduced by Sanger with chain-termination sequencing (90). These methods have dramatically increased in throughput and decreased in cost through the introduction of next-generation massively parallel sequencers (91), and have progressed to single-molecule sequencing platforms (92⇓–94) and CMOS chip-sequencing devices (93⇓–95). The maximal scalability of the present CMOS chip platform provides the potential for further substantial advances in speed and cost. Specifically, for reference, it should be noted that 1 million of the polymerase sensors of Fig. 4 reading bases at accessible Phi29 incorporation rates of 30 to 100 bases per second (73), would generate the quantity of raw sequence data required to read a whole human genome (commonly considered 100 gigabases, or 30x coverage) (91) in under 1 h. CMOS chips with the pixel density of the device shown here (20 μm pitch) (Fig. 1) can have millions of sensors on a large chip, and yet still only cost dollars to produce at high volume. Moreover, the unfettered access to Moore’s Law scaling ensures there can also be a long roadmap of continuous improvements in cost and speed. In the present case, the molecular electronic CMOS sensor scaling limits noted above suggest the long-term potential for chips costing tens of pennies, with the sensor capacity to sequence human genomes in tens of seconds. Indeed, it has been noted (96⇓–98) that extending DNA reading capabilities toward such extreme limits is necessary to make storing digital data in DNA an economical option for the future of zettabyte-scale data storage.
Future Developments.
As the scaling potential of the platform is realized, and the diversity of sensors fully developed, the capacity to read vast amounts of molecular interaction data efficiently and economically has the potential to transform applications, such as whole-genome sequencing, whole-proteome profiling, and DNA data storage, and to make diverse diagnostic assays ever more affordable and widely accessible. Through chips such as these, the 50-year-old vision of molecular electronics as the ultimate means of chip miniaturization may be realized, but driven by their value for advancing the power of sensors, rather than processors (13, 77).
Materials and Methods
More detailed materials and methods are in SI Appendix.
Data Availability
All study data are included in main text and SI Appendix.
Acknowledgments
We thank Sandra Schmid, Eli Yablonovitch, Stan Osher, Stan Nelson, Sue Smalley, and Michael Egholm for helpful discussions and advice, and Mark Reed for providing inspiration and encouragement to pursue Molecular Electronics. This research is supported in part by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), through Contract 2019-19081900003 (development of DNA data-storage reading technology). The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of ODNI, IARPA, or the US government. The US government is authorized to reproduce and distribute reprints for governmental purposes notwithstanding any copyright annotation therein.
Footnotes
Author contributions: C.W.F., H.A., N. Ananthapadmanabhan, T.G., D.A.H., J.R.S., A.G.V., S. Jin, J.M.T., G.M.C., and B.M. designed research; C.W.F., P.S.P., L.A., N. Alagar, N. Ananthapadmanabhan, J.B., C.C., R.D., J.F., C. Garcia, C. Gardner, D.G., D.A.H., G.H., S. Jain, T.J., R.L., Z.M., A.M., P. Mohan, P. Mola, T.N., S.O., Y.O., P.P., N.P., K.P., C.R., T.R., J.R.S., P.S., J.T., A.G.V., S.V., L.Z., P.W.M., and B.M. performed research; J.M.T. and G.M.C. contributed new reagents/analytic tools; C.W.F., P.S.P., S.C., K.J.D., Z.G., A.P.H., Z.M., P. Mudondo, J.M., F.O., D.P., T.S., and B.M. analyzed data; and C.W.F., P.S.P., N. Ananthapadmanabhan, and B.M. wrote the paper.
Reviewers: J.K., University of South Florida College of Arts & Sciences; T.L., Seoul National University; and C.Z., University of Southern California.
Competing interest statement: All authors having the Roswell affiliation (affiliation “a”) are employed by Roswell Biotechnologies, San Diego, CA 92121.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112812119/-/DCSupplemental.
This open access article is distributed under Creative Commons Attribution-NonCommercial-NoDerivatives License 4.0 (CC BY-NC-ND).









